Working with Data
Contents
Working with Data¶
Describing Data using Pandas¶
In this first section, we will use the Pandas package to explore and describe data from O’Connell, K., Berluti, K., Rhoads, S. A., & Marsh, A. A. (2021). Reduced social distancing during the COVID-19 pandemic is associated with antisocial behaviors in an online United States sample. PLoS ONE.
This study assessed whether social distancing behaviors (early in the COVID-19 pandemic) was associated with self-reported antisocial behavior.
# Remember: Python requires you to explictly "import" libraries before their functions are available to use. We will always specify our imports at the beginning of each notebook.
import pandas as pd, numpy as np
Here, we will load a dataset as a pandas.DataFrame, and investigate its attributes. We will see N rows for each subject, and M columns for each variable.
Loading data¶
# here, we are just going to download data from the web
url = 'https://raw.githubusercontent.com/shawnrhoads/gu-psyc-347/master/docs/static/data/OConnell_COVID_MTurk_noPII_post_peerreview.csv'
# load data specified in `filename` into dataframe `df`
df = pd.read_csv(url)
# check type of df
type(df)
pandas.core.frame.DataFrame
# how many rows and columns are in df?
print(df.values.shape) # N x M
(131, 126)
Looks like we will have 131 rows (usually subjects, but can be multiple observations per subject) and 126 columns (usually variables)
# let's output the first 5 rows of the df
print(df.head())
subID mturk_randID suspect_itaysisso Country Region \
0 1001 8797 0 United States CT
1 1002 3756 0 United States IL
2 1003 3798 0 United States OH
3 1004 2965 0 United States TX
4 1005 5953 0 United States NC
ISP loc_US loc_state \
0 AS7015 Comcast Cable Communications, LLC Yes Connecticut
1 AS7018 AT&T Services, Inc. Yes California
2 AS10796 Charter Communications Inc Yes Ohio
3 AS7018 AT&T Services, Inc. Yes Texas
4 AS20115 Charter Communications Yes North Carolina
loc_zipcode loc_County ... education_4yr STAB_total_centered \
0 6511 New Haven County ... 0 -3.946565
1 90280 Los Angeles County ... 0 39.053436
2 44883 Seneca County ... 0 40.053436
3 77019 Harris County ... 1 -9.946565
4 28334 Sampson County ... 0 -17.946566
STAB_total_min32 silhouette_dist_X_min81 silhouette_dist_X_inches \
0 19 441.0 110.332750
1 62 287.0 71.803856
2 63 313.0 78.308731
3 13 452.0 113.084820
4 5 297.0 74.305733
violated_distancing STAB_rulebreak_rmECONOMIC STAB_total_rmECONOMIC \
0 0 9 48
1 1 24 88
2 0 23 85
3 0 8 42
4 0 8 34
STAB_total_rmECONOMIC_centered household_income_coded_centered
0 -2.076336 1.269231
1 37.923664 -3.730769
2 34.923664 -2.730769
3 -8.076336 NaN
4 -16.076336 -2.730769
[5 rows x 126 columns]
Creating custom DataFrames¶
We can also create our own dataframe. For example, here’s a dataframe containing 20 rows and 3 columns of random numbers.
sim_df = pd.DataFrame(np.random.randn(20, 3), index=range(0,20), columns=["column A", "column B", "column C"])
print(sim_df)
column A column B column C
0 0.392950 -1.180451 -0.838785
1 -0.389347 1.017663 -0.424514
2 1.856004 0.285378 -0.120892
3 -0.659527 1.241937 0.754542
4 -2.169041 0.080437 -0.336512
5 1.889987 1.048956 0.740878
6 0.589985 0.581513 0.167113
7 1.844384 -0.015155 -0.285773
8 -1.958567 -0.898125 -0.974897
9 0.005468 -0.030749 -0.672078
10 -0.850622 -1.058297 -0.411733
11 -0.400981 -0.988033 -1.489113
12 1.623723 0.496788 -1.979738
13 1.364699 0.467905 0.228723
14 -0.569220 0.175305 1.545127
15 0.559054 0.083730 -1.018083
16 -0.800119 -0.580347 1.251444
17 0.797440 -0.415382 0.120163
18 -2.143062 0.005168 -0.432682
19 -1.527358 0.585114 -1.219528
We can change the column names using list comprehension
# e.g., change to upper case
sim_df.columns = [x.upper() for x in sim_df.columns]
print(sim_df.head()) #display first 5 rows
COLUMN A COLUMN B COLUMN C
0 0.392950 -1.180451 -0.838785
1 -0.389347 1.017663 -0.424514
2 1.856004 0.285378 -0.120892
3 -0.659527 1.241937 0.754542
4 -2.169041 0.080437 -0.336512
# e.g., change to last element in string
sim_df.columns = [x[-1] for x in sim_df.columns]
print(sim_df.tail()) #display last 5 rows
A B C
15 0.559054 0.083730 -1.018083
16 -0.800119 -0.580347 1.251444
17 0.797440 -0.415382 0.120163
18 -2.143062 0.005168 -0.432682
19 -1.527358 0.585114 -1.219528
Concatenating DataFrames¶
We can concatenate multiple dataframes containing the same columns (e.g., [‘A’,’B’,’C’]) using pd.concat(). This will stack rows across dataframes.
Usage:
pd.concat(
objs,
axis=0,
join="outer",
ignore_index=False,
keys=None,
levels=None,
names=None,
verify_integrity=False,
copy=True,
)
np.zeros((3,3))
array([[0., 0., 0.],
[0., 0., 0.],
[0., 0., 0.]])
# create two new dataframes
# the first will contain only zeros
sim_df1 = pd.DataFrame(np.zeros((3, 3)), index=[20,21,22], columns=["A", "B", "C"])
# the second will contain only ones
sim_df2 = pd.DataFrame(np.ones((3, 3)), index=[23,24,25], columns=["A", "B", "C"])
sim_dfs = [sim_df, sim_df1, sim_df2] # as list of dfs
result = pd.concat(sim_dfs)
print(result)
A B C
0 0.392950 -1.180451 -0.838785
1 -0.389347 1.017663 -0.424514
2 1.856004 0.285378 -0.120892
3 -0.659527 1.241937 0.754542
4 -2.169041 0.080437 -0.336512
5 1.889987 1.048956 0.740878
6 0.589985 0.581513 0.167113
7 1.844384 -0.015155 -0.285773
8 -1.958567 -0.898125 -0.974897
9 0.005468 -0.030749 -0.672078
10 -0.850622 -1.058297 -0.411733
11 -0.400981 -0.988033 -1.489113
12 1.623723 0.496788 -1.979738
13 1.364699 0.467905 0.228723
14 -0.569220 0.175305 1.545127
15 0.559054 0.083730 -1.018083
16 -0.800119 -0.580347 1.251444
17 0.797440 -0.415382 0.120163
18 -2.143062 0.005168 -0.432682
19 -1.527358 0.585114 -1.219528
20 0.000000 0.000000 0.000000
21 0.000000 0.000000 0.000000
22 0.000000 0.000000 0.000000
23 1.000000 1.000000 1.000000
24 1.000000 1.000000 1.000000
25 1.000000 1.000000 1.000000
We can also concatenate using the rows (setting axis=1)
# the first will contain only zeros
sim_df3 = pd.DataFrame(np.zeros((3, 3)), index=[1,2,3], columns=["A", "B", "C"])
# the second will contain only ones
sim_df4 = pd.DataFrame(np.ones((3, 3)), index=[2,4,5], columns=["D", "E", "F"])
result2 = pd.concat([sim_df3, sim_df4], axis=1)
print(result2)
A B C D E F
1 0.0 0.0 0.0 NaN NaN NaN
2 0.0 0.0 0.0 1.0 1.0 1.0
3 0.0 0.0 0.0 NaN NaN NaN
4 NaN NaN NaN 1.0 1.0 1.0
5 NaN NaN NaN 1.0 1.0 1.0
Notice that we have NaNs (not a number) in cells where there were no data (for example, no data in column D for index 1)
We have to be careful because all elements will not merge across rows and columns by default. For example, if the second df also had a column “C”, we will have two “C” columns by default.
# the first will contain only zeros
sim_df5 = pd.DataFrame(np.zeros((3, 3)), index=[1,2,3], columns=["A", "B", "C"])
# the second will contain only ones
sim_df6 = pd.DataFrame(np.ones((3, 4)), index=[2,4,5], columns=["C", "D", "E", "F"])
result3a = pd.concat([sim_df5, sim_df6], axis=1)
print(result3a)
A B C C D E F
1 0.0 0.0 0.0 NaN NaN NaN NaN
2 0.0 0.0 0.0 1.0 1.0 1.0 1.0
3 0.0 0.0 0.0 NaN NaN NaN NaN
4 NaN NaN NaN 1.0 1.0 1.0 1.0
5 NaN NaN NaN 1.0 1.0 1.0 1.0
By default, this method takes the “union” of dataframes. This is useful because it means no information will be lost!
But, now we have two columns named “C”. To fix this, we can have pandas rename columns with matching names using the DataFrame.merge() method.
result3b = sim_df5.merge(sim_df6, how='outer', left_index=True, right_index=True)
print(result3b)
A B C_x C_y D E F
1 0.0 0.0 0.0 NaN NaN NaN NaN
2 0.0 0.0 0.0 1.0 1.0 1.0 1.0
3 0.0 0.0 0.0 NaN NaN NaN NaN
4 NaN NaN NaN 1.0 1.0 1.0 1.0
5 NaN NaN NaN 1.0 1.0 1.0 1.0
We could also take the “intersection” across the two dataframes. We can do that by setting join='inner' (Meaning we only keep the rows that are shared between the two). In the previous case, this would be row ‘2’. All columns would be retained.
result3c = pd.concat([sim_df5, sim_df6], axis=1, join='inner')
print(result3c)
A B C C D E F
2 0.0 0.0 0.0 1.0 1.0 1.0 1.0
Manipulating DataFrames¶
We can also change values within the dataframe using list comprehension.
# first let's view the column "subID"
df['subID']
0 1001
1 1002
2 1003
3 1004
4 1005
...
126 1160
127 1161
128 1162
129 1163
130 1164
Name: subID, Length: 131, dtype: int64
# now, let's change these value by adding a prefix 'sub_' and store in a new column called "subID_2"
df['subID_2'] = ['sub_'+str(x) for x in df['subID']]
df['subID_2']
0 sub_1001
1 sub_1002
2 sub_1003
3 sub_1004
4 sub_1005
...
126 sub_1160
127 sub_1161
128 sub_1162
129 sub_1163
130 sub_1164
Name: subID_2, Length: 131, dtype: object
We can also grab specific elements in the dataframe by specifying rows and columns
print(df['age'][df['subID']==1001])
0 21
Name: age, dtype: int64
# if you know the index (row name), then you can use the `pd.DataFrame.loc` method
df.loc[0,'age']
21
Creating new columns is particularly useful for computing new variables from old variables. For example: for each subject, let’s multiply age by STAB_total.
for index,subject in enumerate(df['subID']):
df.loc[index,'new_col'] = df.loc[index,'age'] * df.loc[index,'STAB_total']
We can extract a column of observations to a numpy array
sub_ids = df['subID'].values
print(sub_ids)
[1001 1002 1003 1004 1005 1007 1011 1013 1014 1015 1016 1017 1021 1022
1023 1024 1026 1028 1029 1032 1033 1034 1035 1040 1043 1044 1045 1047
1048 1049 1051 1053 1054 1056 1057 1058 1059 1060 1061 1062 1063 1064
1066 1068 1069 1070 1071 1072 1073 1074 1075 1076 1077 1078 1079 1080
1082 1083 1084 1085 1086 1087 1088 1089 1090 1091 1092 1093 1094 1095
1096 1098 1099 1100 1101 1102 1104 1105 1107 1108 1109 1110 1111 1112
1113 1114 1115 1116 1117 1118 1119 1120 1121 1122 1123 1124 1125 1126
1127 1128 1129 1130 1131 1132 1133 1134 1135 1136 1137 1138 1139 1141
1144 1145 1146 1147 1148 1149 1150 1151 1152 1154 1155 1156 1157 1158
1160 1161 1162 1163 1164]
print(type(sub_ids))
<class 'numpy.ndarray'>
We can also transpose the dataframe
print(df.T.head())
0 1 2 3 \
subID 1001 1002 1003 1004
mturk_randID 8797 3756 3798 2965
suspect_itaysisso 0 0 0 0
Country United States United States United States United States
Region CT IL OH TX
4 5 6 7 \
subID 1005 1007 1011 1013
mturk_randID 5953 8133 2500 7655
suspect_itaysisso 0 0 0 0
Country United States United States United States United States
Region NC MA FL WI
8 9 ... 121 \
subID 1014 1015 ... 1154
mturk_randID 4616 2318 ... 1123
suspect_itaysisso 0 0 ... 0
Country United States United States ... United States
Region MD LA ... NC
122 123 124 125 \
subID 1155 1156 1157 1158
mturk_randID 2455 7875 6828 3775
suspect_itaysisso 0 0 0 0
Country United States United States United States United States
Region ID CA AZ CO
126 127 128 129 \
subID 1160 1161 1162 1163
mturk_randID 1459 7557 6617 5071
suspect_itaysisso 0 0 0 0
Country United States United States United States United States
Region IL WA CA KY
130
subID 1164
mturk_randID 7031
suspect_itaysisso 0
Country United States
Region MO
[5 rows x 131 columns]
Statistics with DataFrames¶
We can compute all sorts of descriptive statistics on DataFrame columns using the following methods:
count(): Number of non-null observationssum(): Sum of valuesmean(): Mean of valuesmedian(): Median of valuesmode(): Mode of valuesstd(): Standard deviation of valuesmin(): Minimum valuemax(): Maximum valueabs(): Absolute valueprod(): Product of valuescumsum(): Cumulative sumcumprod(): Cumulative product
Here are some examples:
# mean of a column
df["age"].mean()
36.33587786259542
# mean of multiple columns
df[["age","STAB_total"]].mean()
age 36.335878
STAB_total 54.946565
dtype: float64
# median of a column
df["age"].median()
34.0
# compute a summary of metrics on columns
df[["age", "STAB_total"]].describe()
| age | STAB_total | |
|---|---|---|
| count | 131.000000 | 131.000000 |
| mean | 36.335878 | 54.946565 |
| std | 10.075569 | 30.095569 |
| min | 21.000000 | 32.000000 |
| 25% | 29.500000 | 35.000000 |
| 50% | 34.000000 | 42.000000 |
| 75% | 41.000000 | 55.500000 |
| max | 65.000000 | 139.000000 |
# group means by sex
df.groupby("sex")[["age", "STAB_total"]].mean()
| age | STAB_total | |
|---|---|---|
| sex | ||
| Female | 38.264151 | 49.792453 |
| Male | 35.025641 | 58.448718 |
# group means by sex and education
df.groupby(["sex","education_coded"])[["age", "STAB_total"]].mean()
| age | STAB_total | ||
|---|---|---|---|
| sex | education_coded | ||
| Female | Four-year college degree (B.A., B.S.) | 37.592593 | 54.074074 |
| High school graduate or G.E.D. | 42.400000 | 37.600000 | |
| Master's degree (for example, M.B.A., M.A., M.S.) | 38.200000 | 55.800000 | |
| Some college | 38.875000 | 48.875000 | |
| Two-year college degree (A.A., A.S.) | 37.375000 | 40.125000 | |
| Male | Doctoral or professional degree (for example, Ph.D., M.D., J.D., D.V.M., D.D.M) | 31.500000 | 34.500000 |
| Four-year college degree (B.A., B.S.) | 35.125000 | 64.000000 | |
| High school graduate or G.E.D. | 31.777778 | 63.888889 | |
| Master's degree (for example, M.B.A., M.A., M.S.) | 39.000000 | 77.375000 | |
| Some college | 34.368421 | 48.157895 | |
| Two-year college degree (A.A., A.S.) | 36.750000 | 41.625000 |
# group counts by sex and education
df.groupby(["sex","education_coded"])[["age", "STAB_total"]].count()
| age | STAB_total | ||
|---|---|---|---|
| sex | education_coded | ||
| Female | Four-year college degree (B.A., B.S.) | 27 | 27 |
| High school graduate or G.E.D. | 5 | 5 | |
| Master's degree (for example, M.B.A., M.A., M.S.) | 5 | 5 | |
| Some college | 8 | 8 | |
| Two-year college degree (A.A., A.S.) | 8 | 8 | |
| Male | Doctoral or professional degree (for example, Ph.D., M.D., J.D., D.V.M., D.D.M) | 2 | 2 |
| Four-year college degree (B.A., B.S.) | 32 | 32 | |
| High school graduate or G.E.D. | 9 | 9 | |
| Master's degree (for example, M.B.A., M.A., M.S.) | 8 | 8 | |
| Some college | 19 | 19 | |
| Two-year college degree (A.A., A.S.) | 8 | 8 |
We can also correlate 2 or more variables
df[["age","STAB_total","socialdistancing"]].corr(method="spearman")
| age | STAB_total | socialdistancing | |
|---|---|---|---|
| age | 1.000000 | -0.145465 | 0.007075 |
| STAB_total | -0.145465 | 1.000000 | -0.335639 |
| socialdistancing | 0.007075 | -0.335639 | 1.000000 |
Visualizing Data using Matplotlib¶
To understand what our data look like, we will visualize it in different ways.
import matplotlib.pyplot as plt
%matplotlib inline
Let’s plot the distribiton of one variable in our data
plt.hist(df['age'], bins=9)
plt.xlabel("Age")
plt.ylabel("Number of Subjects")
Text(0, 0.5, 'Number of Subjects')
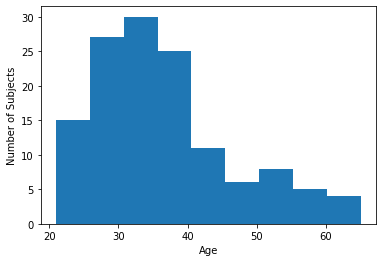
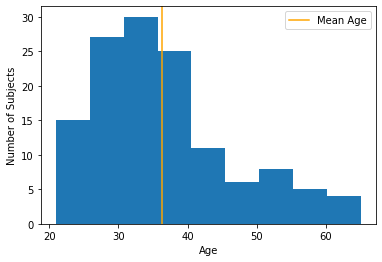
Let’s see what percentage of subjects have a below-average score:
mean_age = np.mean(df['age'])
frac_below_mean = (df['age'] < mean_age).mean()
print(f"{frac_below_mean:2.1%} of subjects are below the mean")
63.4% of subjects are below the mean
We can also see this by adding the average score to the histogram plot:
plt.hist(df['age'], bins=9)
plt.xlabel("Age")
plt.ylabel("Number of Subjects")
plt.axvline(mean_age, color="orange", label="Mean Age")
plt.legend();
plt.show();
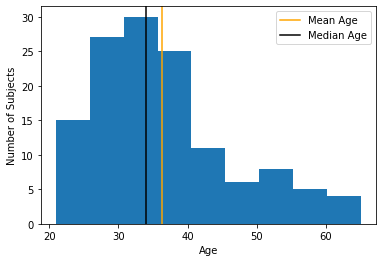
Comparing mean and median
med_age = np.median(df['age'])
plt.hist(df['age'], bins=9)
plt.xlabel("Age")
plt.ylabel("Number of Subjects")
plt.axvline(mean_age, color="orange", label="Mean Age")
plt.axvline(med_age, color="black", label="Median Age")
plt.legend();
plt.show();