Linear Modeling
Contents
# If you are using Anconda, please re-download this file to current directory:
# https://raw.githubusercontent.com/shawnrhoads/gu-psyc-347/master/course-env.yml
# Then, run this cell
# !conda env update --file course-env.yml
Linear Modeling¶
This tutorial was inspired by and adapted from the Neuromatch Academy tutorials [CC BY 4.0].
Goals of this tutorial¶
What is a linear model?¶
In its most basic form, a linear model can be written like this (e.g., a simple linear regression):
where \(y\) is the dependent (outcome) variable and \(x\) is an independent (explanatory/manipulated) variable—these variables represent data (each participant \(n\) has an observation at \(x\), \(y\)).
\(y\) is a function of \(x\) and is determined by 2 components:
a non-random component: \(intercept + \beta*x_n\)
random component: \(\epsilon_n\)
The \(intercept\) is the value of \(y\) when \(x=0\). \(\beta\) is a weighted parameter that determines the slope of the fitted linear model. We will determine the values of these parameters by fitting a linear model.
The \(error\) (\(\epsilon_n\)) describes the random component of the linear relationship between \(x\) and \(y\)—this is the difference between the true values (\(y\)) and the predicted values (\(\hat{y}\)). We can solve this equation for the error:
# let's begin by importing packages
import pandas as pd, numpy as np, requests
from scipy.stats import truncnorm
from scipy.optimize import minimize
import matplotlib.pyplot as plt
from matplotlib.patches import Rectangle
import ipywidgets as widgets
import seaborn as sns
%matplotlib inline
Model Simulations¶
Simulations are great ways to test models. By creating a simple synthetic dataset, we will know the true underlying model which allows us to see how our estimation efforts compare in uncovering the real model.
Below, we will simulate a linear relationship between two variables x (antisocial behavior) and y (distance kept from others during the COVID-19 pandemic), and then we will add some “noise” to those data. This will help us gain a better understanding about expected results from O’Connell, et al., (2021).
# setting a fixed seed to our random number generator
# ensures we will always
# get the same psuedorandom number sequence
np.random.seed(2021)
# Let's set some parameters
beta = -1.75
n_subjects = 131
# Draw x
intercept = 347
x0 = np.ones((n_subjects, 1))
lower, upper = 32, 139 # range of STAB scores
mu, sigma = 54.9, 30.1 # mean, and standard deviation
x1 = truncnorm.rvs((lower-mu)/sigma,
(upper-mu)/sigma,
loc=mu,
scale=sigma,
size=(n_subjects,1))
X = np.hstack((x0, x1))
# sample from a standard normal distribution
noise = np.random.normal(2, 10, (n_subjects,1))
# calculate y
y = intercept + beta * x1 + noise
# Plot a histogram
fig, ax = plt.subplots()
ax.hist(x1)
ax.set(xlabel='Antisocial behavior', ylabel='Frequency')
plt.show()

# Plot a histogram
fig, ax = plt.subplots()
ax.hist(y)
ax.set(xlabel='Distance from others', ylabel='Frequency')
plt.show()
# Plot the results
fig, ax = plt.subplots()
ax.scatter(x1, y) # produces a scatter plot
ax.set(xlabel='Antisocial behavior', ylabel='Distance from others')
plt.show()
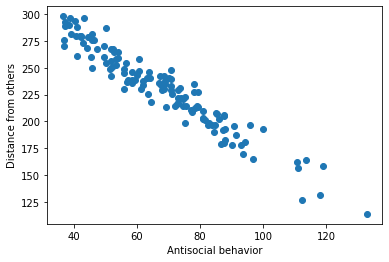
Now that we have our noisy dataset, we can try to estimate the underlying model that produced it. We use MSE to evaluate how successful a particular slope estimate \(\hat{\beta}\) is for explaining the data, with the closer to 0 the MSE is, the better our estimate fits the data.
Model Fitting¶
Now, we will fit our simulated data to a simple linear regression, using least-squares optimization.
Ordinary least squares is a very common optimization procedure that we are going to use for data fitting.
Let’s recall our simple linear model above. Suppose we have a set of measurements, \(y_{n}\) (the “dependent” variable) obtained for different input values, \(x_{n}\) (the “independent” or “explanatory” variable). Suppose we believe the measurements are proportional to the input values, but are corrupted by some (random) measurement errors, \(\epsilon_{n}\), that is:
for some unknown parameters (\(\beta_0\), \(\beta_1\)). The least squares regression problem uses mean squared error (MSE) as its objective function, it aims to find the values of \(\beta_0\) and \(\beta_1\) by minimizing the average of squared errors:
We will now explore how MSE is used in fitting a linear regression model to data.
Computing MSE¶
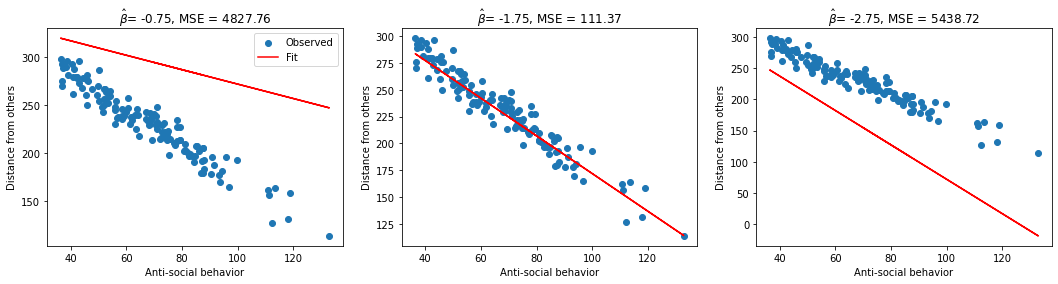
In this exercise we will implement a method to compute the mean squared error (MSE) for a set of inputs \(x\), measurements \(y\), and slope estimates \(\hat{\beta_0}\) and \(\hat{\beta_1}\). We will then compute and print the MSE for 3 different estimates of \(\hat{\beta_1}\).
def mse(beta_hats, X, y):
"""Compute the mean squared error
Args:
beta_hats (list of floats): A list containing the estimate of the intercept parameter and the estimate of the slope parameter
X (ndarray): An array of shape (samples,) that contains the input values.
y (ndarray): An array of shape (samples,) that contains the corresponding measurement values to the inputs.
Returns:
y_hat (float): The estimated y_hat computed from X and the estimated parameter(s).
mse (float): The mean squared error of the model with the estimated parameter(s).
"""
assert len(beta_hats) == np.shape(X)[1]
# Compute the estimated y
y_hat = 0
for index, b in enumerate(beta_hats):
y_hat += b*X[:,index]
y_hat = y_hat.reshape(len(y_hat),1)
# Compute mean squared error
mse = np.mean((y - y_hat)**2)
return y_hat, mse
intercept = 347
possible_betas = [-0.75, -1.75, -2.75]
for beta_hat in possible_betas:
y_hat, MSE_val = mse([intercept, beta_hat], X, y)
print(f"beta_hat of {beta_hat} has an MSE of {MSE_val:.2f}")
beta_hat of -0.75 has an MSE of 4827.76
beta_hat of -1.75 has an MSE of 111.37
beta_hat of -2.75 has an MSE of 5438.72
fig, axes = plt.subplots(ncols=3, figsize=(18, 4))
for beta_hat, ax in zip(possible_betas, axes):
# True data
ax.scatter(x1, y, label='Observed') # our data scatter plot
# Compute and plot predictions
y_hat, MSE_val = mse([intercept, beta_hat], X, y)
ax.plot(x1, y_hat, color='r', label='Fit') # our estimated model
ax.set(
title= fr'$\hat{{\beta}}$= {beta_hat}, MSE = {MSE_val:.2f}',
xlabel='Anti-social behavior',
ylabel='Distance from others')
axes[0].legend()
plt.show()
Model parameters¶
Using an interactive widget, we can easily see how changing intercept and slope estimates change a model fit. We display the residuals (differences between observed and predicted data) as line segments between the actual data and the corresponding predicted data on the model fit line.
# interactive display
import matplotlib.pyplot as plt
import ipywidgets as widgets
import numpy as np
# set up data
x0_viz = np.ones((131, 1))
x1_norm = np.random.normal(loc=54.9,scale=30.1,size=(131,1))
x1_viz = x1_norm - min(x1_norm)
X_viz = np.hstack((x0_viz, x1_viz))
noise_viz = np.random.normal(2, 10, (131,1))
y_viz = 347 - 1.75 * x1_viz + noise_viz
@widgets.interact(beta_hat=widgets.FloatSlider(-1.75, min=-3, max=0),
intercept=widgets.FloatSlider(347, min=100, max=400))
def plot_data_estimate(intercept, beta_hat):
# compute error
X, y = X_viz, y_viz
y_hat = (intercept*X[:,0] + beta_hat*X[:,1]).reshape(len(y),1)
MSE_val = np.mean((y - y_hat)**2)
# plot
fig, ax = plt.subplots()
ax.scatter(X[:,1], y, label='Observed') # our data scatter plot
ax.plot(X[:,1], y_hat, color='r', label='Fit') # our estimated model
# plot residuals
ymin = np.minimum(y, y_hat)
ymax = np.maximum(y, y_hat)
ax.vlines(X[:,1], ymin, ymax, 'g', alpha=0.5, label='Residuals')
ax.set(
title=fr"$intercept={intercept:0.2f}, \hat{{\beta}}$ = {beta_hat:0.2f}, MSE = {MSE_val:.2f}",
xlabel='x',
ylabel='y')
ax.legend()
plt.show()
# interactive display (if not using Jupyter Book)
%config InlineBackend.figure_format = 'retina'
@widgets.interact(beta_hat=widgets.FloatSlider(-1.75, min=-3, max=0),
intercept=widgets.FloatSlider(347, min=100, max=400))
def plot_data_estimate(intercept, beta_hat):
# compute error
y_hat = (intercept*X[:,0] + beta_hat*X[:,1]).reshape(len(y),1)
MSE_val = np.mean((y - y_hat)**2)
# plot
fig, ax = plt.subplots()
ax.scatter(X[:,1], y, label='Observed') # our data scatter plot
ax.plot(X[:,1], y_hat, color='r', label='Fit') # our estimated model
# plot residuals
ymin = np.minimum(y, y_hat)
ymax = np.maximum(y, y_hat)
ax.vlines(X[:,1], ymin, ymax, 'g', alpha=0.5, label='Residuals')
ax.set(
title=fr"$intercept={intercept:0.2f}, \hat{{\beta}}$ = {beta_hat:0.2f}, MSE = {MSE_val:.2f}",
xlabel='x',
ylabel='y')
ax.legend()
plt.show()
Least-squares minimization¶
While the approach detailed above (computing MSE at various values of \(\hat\beta\)) quickly got us to a good estimate, it still relied on us guessing which beta values to select. If we didn’t pick good guesses to begin with, we might miss the best possible parameter values.
Thus, there must be a better way “guess-timate” them, right?
Why don’t we try an exhaustive search of across a specified range of parameter values?
# Exhaustive search
param_b0 = np.arange(325, 375, 1)
param_b1 = np.arange(-3., 0., .25)
first_run = 'first'
mse_out = []
for b0 in param_b0:
for b1 in param_b1:
if first_run is not None:
first_run = None
Params = [b0, b1]
else:
Params = np.vstack((Params, [b0, b1]))
mse_out = np.append(mse_out, mse([b0, b1], X, y)[1])
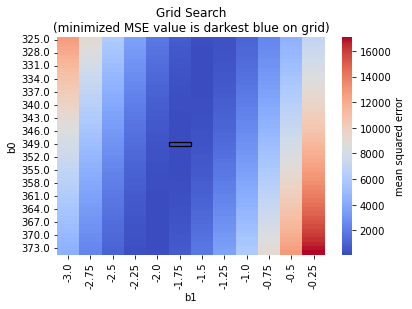
# From out exhaustive search:
# let's see if we can recover the index with the smallest MSE
min_index = np.argmin(mse_out)
print(f'b0, b1 = {Params[min_index]}, MSE = {mse_out[min_index]:.2f}')
b0, b1 = [349. -1.75], MSE = 105.48
We see that our fit is \(b_0 = 349\) and \(b_1 = -1.75\), which is quite close to our original!
We can also plot our search onto a heatmap!
grid_search_data = pd.DataFrame({'b0': Params[:,0],
'b1': Params[:,1],
'MSE': mse_out})
data_pivoted = grid_search_data.pivot("b0", "b1", "MSE")
ax = sns.heatmap(data_pivoted,
cmap='coolwarm',
cbar_kws={'label': 'mean squared error'})
# add rectangle around minimum
ax.add_patch(Rectangle((5, 24), 1, 1, fill=False, edgecolor='black', lw=1.25))
plt.title("Grid Search\n(minimized MSE value is darkest blue on grid)")
plt.show()
scipy.optimize.minimize¶
But, writing an exhaustive search by hand is prone to errors, too! It depends largely on the specified parameters for the search (i.e., we only sampled 300 to 450 in increments of .5 above for \(b_0\)).
It can also be very time-consuming and computationally-expensive.
Instead, we can utilize minimization algorithms that we optimized to solve minimization problems like this. Let’s use the scipy.optimize.minimize function, which takes in the following inputs:
a cost function to minimize (
mse()in our case)starting points for the parameters to estimate (starting points for
b0andb1in our case)other arguments that need to be input into our objective function (
Xandyin our case)
It will return an OptimizeResult object containing the estimated parameters (along with the value of minimized MSE)
def mse_to_minimize(beta_hats, X, y):
"""Compute the mean squared error
Args:
beta_hats (list of floats): A list containing the estimate of the intercept parameter and the estimate of the slope parameter
X (ndarray): An array of shape (samples,) that contains the input values.
y (ndarray): An array of shape (samples,) that contains the corresponding measurement values to the inputs.
Returns:
mse (float): The mean squared error of the model with the estimated parameter(s).
"""
assert len(beta_hats) == np.shape(X)[1]
# Compute the estimated y
y_hat = 0
for index, b in enumerate(beta_hats):
y_hat += b*X[:,index]
y_hat = y_hat.reshape(len(y_hat),1)
# Compute mean squared error
mse = np.mean((y - y_hat)**2)
return mse
def plot_observed_vs_predicted(beta_hats, X, y):
""" Plot observed vs predicted data
Args:
x (ndarray): observed x values
y (ndarray): observed y values
y_hat (ndarray): predicted y values
beta_hats (ndarray): An array of shape (betas,) that contains the estimate of the slope parameter(s)
"""
y_hat, MSE_val = mse(beta_hats, X, y)
fig, ax = plt.subplots()
ax.scatter(X[:,1], y, label='Observed') # our data scatter plot
ax.plot(X[:,1], y_hat, color='r', label='Fit') # our estimated model
# plot residuals
ymin = np.minimum(y, y_hat)
ymax = np.maximum(y, y_hat)
ax.vlines(X[:,1], ymin, ymax, 'g', alpha=0.5, label='Residuals')
ax.set(
title=fr"$intercept={beta_hats[0]:0.2f}, \hat{{\beta}}$ = {beta_hats[1]:0.2f}, MSE = {MSE_val:.2f}",
xlabel='x',
ylabel='y')
ax.legend()
plt.show()
# minimize MSE using scipy.optimize.minimize
res = minimize(mse_to_minimize, # objective function
(350, -1.75), # estimated starting points
args=(X, y)) # arguments
print(f'b0, b1 = {(res.x)}, MSE = {res.fun}')
plot_observed_vs_predicted([res.x[0], res.x[1]], X, y)
b0, b1 = [352.01054797 -1.78736471], MSE = 104.68617090696347
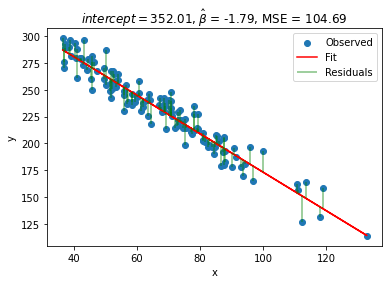
We see that we can get a better fit: \(b_0 = 352.01\) and \(b_1 = -1.79\), which fairly accurate! Note that our MSE value is also smaller than last time!
Analytic solution for Least Squares Optimization (Bonus)¶
While the approach detailed above (computing MSE at various values of \(\hat\beta\)) got us to a good estimate, we still had to rely on evaluating the MSE value across a grid of hand-specified values.
If we don’t pick a good range to begin with, or with enough granularity, we might miss the best possible parameters. Let’s go one step further, and instead of finding the minimum MSE from a set of candidate estimates, let’s solve for it analytically.
We can do this by minimizing a “cost function”. Mean squared error is a convex objective function, therefore we can compute its minimum using calculus (see appendix below for this derivation)! After computing the minimum, we find that:
This is known as solving the normal equation. For different ways of obtaining the solution, see the notes on Least Squares Optimization by Eero Simoncelli.
def ordinary_least_squares(X, y):
"""Ordinary least squares estimator for linear regression.
Args:
X (ndarray): design matrix of shape (n_samples, n_regressors)
y (ndarray): vector of measurements of shape (n_samples)
Returns:
ndarray: estimated parameter values of shape (n_regressors)
"""
# Compute beta_hat using OLS
beta_hat = np.linalg.inv(X.T @ X) @ X.T @ y
return beta_hat
# Compute beta_hat
beta_hat2 = ordinary_least_squares(X, y)
print(f"b0={beta_hat2[0]}, b1={beta_hat2[1]}")
# Compute MSE
y_hat2 = X @ beta_hat2
print(f"MSE = {np.mean((y - y_hat2)**2):.2f}")
b0=[352.0105366], b1=[-1.78736455]
MSE = 104.69
Actual data versus fitted data¶
In addition to reviewing the residuals, we can also look at the actual simulated data (y or \(y\)) versus fitted data (y_hat or \(\hat{y}\)). In other words, we can visualize the how the fitted values (\(\hat{y}\)) compare to the actual data (\(y\)) to assess how well our model recovered our original parameters. We can also correlate \(y\) with \(\hat{y}\) (a good model fit will show a strong correlation). This is helpful because we are often dealing with more than one dimension.
# correlate y and y_hat
corrcoef = np.corrcoef(y.flatten(),y_hat2.flatten())[0,1]
# Plot the results
fig, ax = plt.subplots()
ax.scatter(y, y_hat2) # produces a scatter plot
ax.set(xlabel='y (simulated data + noise)', ylabel=fr'$\haty$ (fitted data)')
plt.title((f'r={corrcoef:.2f}'))
plt.show()
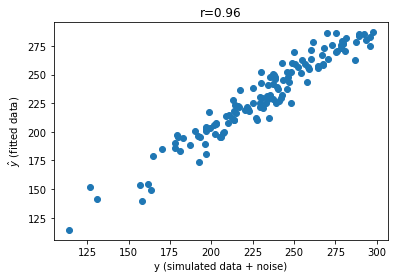
Awesome! Looks pretty nice!
Comparison to a statistics package¶
Okay, cool!
Let’s see how our manual linear model fitting compares to a typical statistical packages. Let’s import scipy.stats and run a simple linear regression with our observed data!
from scipy import stats
stat_res = stats.linregress(X[:,1],y[:,0])
y_hat_new = (stat_res.intercept + stat_res.slope*X[:,1]).reshape((len(y),1))
print(f'b1 = {stat_res.slope:.2f} , b0 = {stat_res.intercept:.2f}, MSE = {np.mean((y - y_hat_new)**2):.2f}')
b1 = -1.79 , b0 = 352.01, MSE = 104.69
Multivariate data¶
Now that we have considered the univariate case, we turn to the general linear model case, where we can have more than one regressor, or feature, in our input.
Recall that our original univariate linear model was given as
where \(\beta\) is the slope and \(\epsilon\) some noise. We can easily extend this to the multivariate scenario by adding another parameter for each additional feature
where \(\beta_0\) is the intercept and \(d\) is the number of features (it is also the dimensionality of our input).
For now, we will focus on the two-dimensional case (\(d=2\)), which allows us to easily visualize our results.
In this case our model can be writen as:
# Let's add age
np.random.seed(2021)
b1 = -1.84
b2 = -.68
lower2, upper2 = 21, 65 # range of ages
mu2, sigma2 = 36.34, 10.08 # mean and standard deviation
x2 = np.random.normal(mu2, sigma2,
size=(n_subjects,1))
X_a = np.hstack((x0, x1, x2))
# sample from a standard normal distribution
noise = np.random.normal(2, 10, (n_subjects,1))
y_a = (b0 + b1*x1 + b2*x2 + noise).reshape((n_subjects,1))
# Compute beta_hat
beta_hat3 = ordinary_least_squares(X_a, y_a)
print(f"b0={beta_hat3[0]}, b1={beta_hat3[1]}, b2={beta_hat3[2]}")
# Compute MSE
y_hat3 = (X_a @ beta_hat3)
print(f"MSE = {np.mean((y_a - y_hat3)**2):.2f}")
b0=[374.49097358], b1=[-1.81811949], b2=[-0.6804654]
MSE = 101.97
Summary¶
Ordinary least squares regression is an optimization procedure that can be used for fitting data to models (i.e., predicting a value for \(y\) given \(x\) by minimizing the MSE)
Note: In this case, there is also an analytical solution to the minimization problem (see below). But as models become more complex, we will need to use a minimization algorithm to help us out!
Appendix: Least-squares derivation¶
Here’s the derivation of the least squares solution.
First, set the derivative of the error with respect to \(\beta\) equal to zero (use chain rule),
Now solving for \(\beta\), we obtain an optimal value of:
This can be written in vector notation as:
This is equivalent to the following code (used above):
beta_hat = np.linalg.inv(X.T @ X) @ X.T @ y
This is known as solving the normal equations. For different ways of obtaining the solution, see the notes on Least Squares Optimization by Eero Simoncelli.
